An excellent way to watch proteins fold is to probe the inside of a microfluidics device with light. This tiny device contains micron-sized three-dimensional (3D) transparent channels that carry small amounts of liquid. Inside the channels, the fluid flow is laminar, i.e., there is no turbulence. Consequently, fluid flow through them is predictable and easily modeled. Microfluidics devices have been used to study chemical reaction kinetics and control chemical and biological reactions.
Research associate Emily Gibson, Fellow Ralph Jimenez, and colleagues from the Colorado School of Mines recently used a home-built scanning two-photon optical microscope to study the 3D mixing of two solutions in a microfluidic device. The researchers were able to create 3D maps of the concentrations of one of the dissolved chemicals during mixing by using a fluorescent dye to probe the mixing dynamics. In so doing, they showed it was possible to determine precisely how fast mixing occurred and exactly where mixing took place with respect to the mixing junction. This straightforward technique will lead to more accurate measurements of kinetics when using microfluidic mixers to trigger chemical reactions.
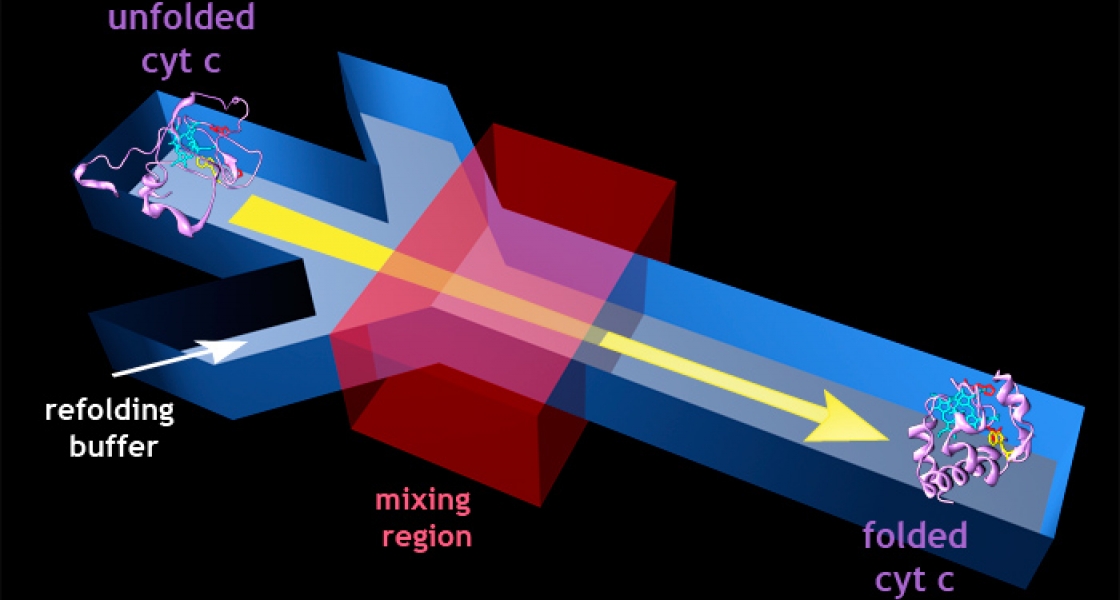
This experiment set the stage for using microfluidics devices in conjunction with spectroscopic techniques to investigate the folding of cytochrome c, an important protein in transforming energy for use in cells. Gibson and her colleagues started with unfolded cytochrome c molecules, which had been dissolved in a concentrated salt solution. In the unfolded state, these proteins have an amino acid residue that fluoresces when excited by ultraviolet light.
To perform the folding experiment, the high-salt protein solution was sent down the channels of the microfluidics device and diluted with buffer introduced at a mixing junction. As the salt concentration fell, the researchers monitored changes in fluorescence to "watch" the cytochrome c molecules fold into their natural 3D biological configuration. As the natural configuration took shape, the fluorescing amino acid group ended up close to the protein’s cofactor, quenching the fluorescence. By closely tracking the changes in fluorescence, the researchers were able to measure the dynamics of protein folding.
Next the researchers plan to use ultrafast laser spectroscopy to monitor changes in the motion and flexibility of the molecule’s amino acid chains during the folding process. - Julie Phillips