Our lives depend on heme. As part of hemoglobin, it carries oxygen to our tissues. As part of cytochrome c, it helps transform the energy in food into the energy-rich molecule ATP (adenosine triphosphate) that powers biochemical reactions that keep us alive and moving. As part of cytochrome P450, it helps break down toxic chemicals in our bodies.
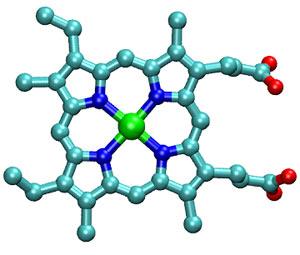
What is this thing called heme? And, how does it do such amazing work inside our bodies? Scientists know that heme is a large ringed molecule called a porphyrin that has an iron atom sitting in the middle of it. In the heme molecule shown at right, the iron atom is green and the atoms in the porphyrin ring are carbon (teal), nitrogen (dark blue), hydrogen (not shown), and oxygen (red). In the figure below, the heme is embedded in (and bonded to) cytochrome c.
Understanding how heme functions at the biomolecular level is a hot research topic for biophysicists, including Research Associate Byung Moon Cho, Graduate Student Fredrik Carlsson, and Associate Fellow Ralph Jimenez. The JILA researchers use photon echo spectroscopy to study the motions of proteins associated with or attached to heme groups. Understanding these motions will help scientists figure out how heme proteins accomplish their important work inside our cells.
Recently, the JILA scientists began a series of studies to measure and quantify the motions of cytochrome c and two other zinc-containing porphyrins. To allow them to make the most precise measurements possible, they substituted a zinc atom for the iron atom in the cytochrome c. To determine which motions are important biologically, the researchers compared the motions of natural cytochrome c, which has an active, three-dimensional structure, with the motions of identical, but denatured proteins, which have lost both their 3D structure and biological activity. Their spectroscopic technique clearly differentiated between active, functional cytochrome c and its denatured, nonfunctional cousin. It also provided information on the protein's binding with substrates, interaction with solvents, and energy states.
In the future, the researchers would like to get sufficient information about the specific motions of cytochrome c to be able to relate them to the changes in entropy that drive the biochemical reactions mediated by this vital protein.
An article describing this work has been accepted for publication in the Journal of Chemical Physics. - Julie Phillips