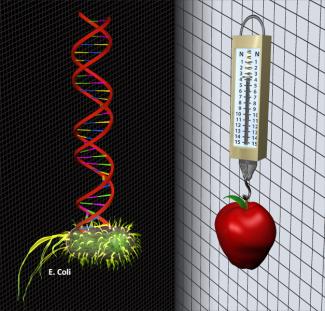
The Perkins group is helping to develop DNA as a force standard for the nano world. Polymers of DNA act like springs, and DNA's elasticity may one day provide a force standard from 0.1–10 piconewtons (pN). One pN is the force exerted when 1 mW of light reflects off a mirror or the approximate weight of one hundred E. coli cells. DNA is an excellent candidate for a force standard because its double helix is reproduced with exquisite fidelity, which allows researchers (or cells) to build it with atomic precision.
DNA-based force standards would be a huge improvement over the current method of estimating tiny forces with measurements of the torque transfer between one atomic force microscope (AFM) tip pushing against a second AFM tip. This method is very hard to do, very slow, and will never be able to provide the precision possible with a biomolecule. "Biomolecules are going to be great precision measurement tools," Perkins says.
The current challenge for Fellow Tom Perkins, research associate Yeonee Seol, and their theoretical colleagues from the Universities of Colorado and Pennsylvania is to determine how to precisely measure the elasticity of relatively short strands of DNA. Until they tackled the problem, elasticity measurements of short (~ 630 nm) lengths of DNA had systematic error rates of up to 18% — which is absolutely abysmal for any kind of precision measurement!
The Perkins group determined that the biggest problem was not with elasticity measurements of double-stranded DNA, but rather with the theoretical model describing DNA elasticity. The old theory, known as the worm-like chain (WLC) model, neglected to consider three important effects that influence elasticity: (1) the finite length of the DNA chain, (2) rotation of the bead attached to the DNA during elasticity measurements, and (3) boundary conditions. For example, the Perkins group experimentally showed that the assumption that DNA polymers were infinitely long only works well for very large molecules. In the 300–2000 nm length range used in most research, this faulty assumption leads to significant errors in interpreting experimental data.
To correct this problem, the Perkins group teamed up with its theorist colleagues to incorporate these three corrections into the WLC, creating the finite worm-like chain (FWLC) solution. In so doing, they reduced the systematic error rate of elasticity measurements by threefold. The new FWLC reproduces experimental reality much more closely than its predecessor did.
By elucidating the origin of a significant error in interpreting DNA elasticity, the group has furthered the adoption of DNA as a molecular force standard. In addition, experimentalists now have a better model for analyzing single molecule experiments, enabling accurate and precise experiments with shorter DNA polymers. For instance, the new model has already explained why two of the nation’s top groups, Steve Block’s group at Stanford and Carlos Bustamante’s group at the University of California at Berkeley, had consistently found significantly different values of DNA elasticity: The two groups had performed their experiments on DNA polymers of different lengths. Now these groups (and others) can use the new model to guide future research in nucleic acid-enzyme interactions.
There’s still work to be done before DNA-based force standards are ready for prime time, however. Researchers must figure out how to further reduce systematic error rates of DNA elasticity measurements to 1% or less. Because of the Perkins group’s improved theoretical model validated with experimental data, researchers have a good idea where to begin — and how to better evaluate what they discover. - Julie Phillips